-
Notifications
You must be signed in to change notification settings - Fork 528
Description
When calling the unbalanced barycenter function without going straight to the right method, the weights do not get passed on in the if-cases lines 1003 and following in unbalanced.py.
I found this by running the example script plot_UOT_barycenter_1D.py and changing the function call in there as mentioned in another issue raised earlier, changing
weights
to
weights=weights
. The interpolation supposed to happen only generates the step with the weights 0.5 and 0.5 as per default in unbalanced.barycenter_unbalanced_sinkhorn. The following image shows the result of the interpolation.
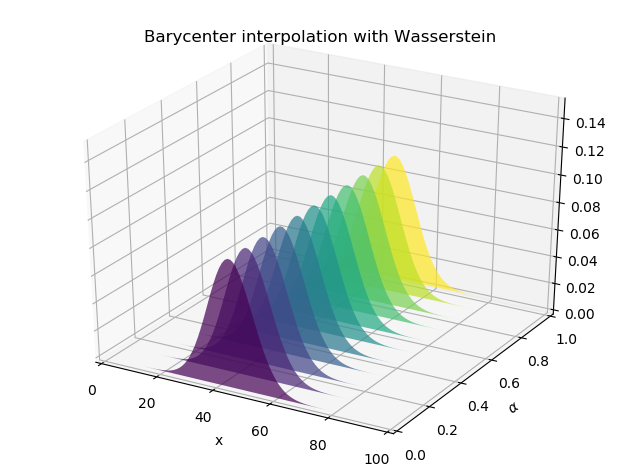
By adding
weights=weights
into the calls in the if-cases at the bottom of unbalanced.py the interpolation works, although there are some numerical problems then, which I do not understand OT well enough yet to judge. The following plot shows all steps from the third, which is the first without any nan in it.
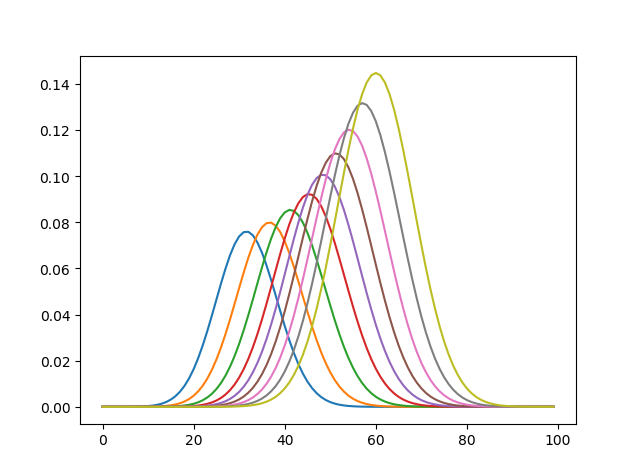
Now it is showing changes in the distributions from one interpolation step to the next.
My system is
Windows-10-10.0.18362-SP0
Python 3.7.6 (default, Jan 8 2020, 20:23:39) [MSC v.1916 64 bit (AMD64)]
NumPy 1.18.1
SciPy 1.4.1
POT 0.6.0